
Monitoring fish communities is vital for river conservation.
This study used eDNA metabarcoding to track fish populations across three French rivers, revealing seasonal and hydrological influences. Unlike electrofishing, eDNA provided a more sensitive biodiversity snapshot, detecting species more consistently in stable rivers while revealing sporadic occurrences in a lake-influenced river.
The findings prove eDNA sampling, enhancing its effectiveness for large-scale monitoring.
Why this matters?
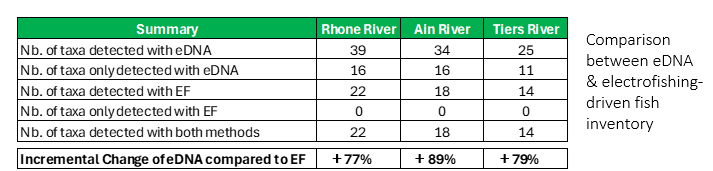
This study demonstrated that eDNA can detect up to 89% more species than electrofishing (EF) and also showed how seasonal and hydrological factors influence detection, guiding researchers toward optimized sampling strategies.
By improving the accuracy and efficiency of biodiversity assessments, eDNA offers a non-invasive, cost-effective alternative to traditional methods, making it a game-changer for freshwater conservation and management.
Results were published and can be accessed here.

Location: the three sampling sites were located in eastern France, at the Rhone River, Ain River, Tiers River.
Ecosystem: Freshwater River.
Depth: Surface Sampling.
Sampling method: each of the 3 stations was sampled at two-month intervals, using three 30-liter filtrations on the left bank, right bank, and in the middle of the watercourse.
Taxonomic group: fishes, using teleo primer.

