
The accelerating impact of global change threatens biodiversity worldwide, with coral reef fishes among the most affected. Safeguarding the biodiversity of these fishes requires advanced monitoring to better understand coral reef dynamics.
Recently, the comparison of of eDNA metabarcoding and extensive visual censuses has unveiled groundbreaking insights into the spatial organization of these incredibly rich marine ecosystems.
Why this matters?
The study highlights the application of environmental DNA (eDNA) metabarcoding to study fish biodiversity on coral reefs, revealing critical insights into these ecosystems.
- Enhanced biodiversity detection: eDNA detected 16% more fish taxa than traditional visual surveys, uncovering hidden patterns, particularly in pelagic, reef-associated, and crypto-benthic species.
- Spatial patterns and regional diversity: The study confirmed known biodiversity gradients (e.g., Coral Triangle as the richest region) while identifying strong regional differentiation and localized biodiversity turnover.
- Complementary methodologies: While eDNA excels in identifying elusive or transient species, visual surveys remain crucial for capturing taxa less represented in genetic reference databases.
Results were published in the journal Proceedings of the Royal Society B and can be accessed here.
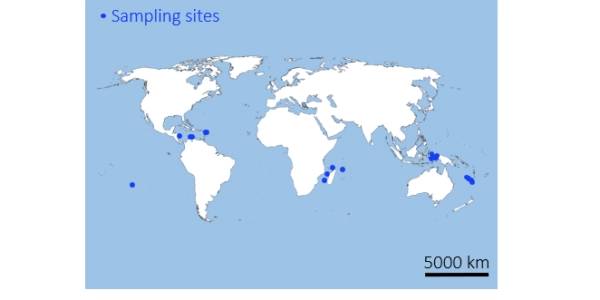
Location: Global
Ecosystem: Coastal (coral reefs)
Depth: 0-40 m
Sampling method: via 30 liter filters on 72 non-overlapping 2 km surface transects (using peristaltic pump on a ship) and 28 2 l filters at bottom fixed points (via scuba diving)
Taxonomic group: fishes, using teleo primers.

