
Large rivers like the Rhône are vital ecosystems, yet monitoring their fish populations is costly and challenging.
Traditional methods like electrofishing are limited, but eDNA offers a broader reach.
Here, by analyzing genetic traces in water samples collected along 500 km of the Rhône River, eDNA accurately capture fish assemblages, reflecting both species presence and relative abundance.
Why this matters?
Aquatic systems, especially rivers, are recognized as highly degraded in terms of both species diversity and ecological functioning. The rate of decline in aquatic populations is almost twice that of terrestrial populations. Given this dramatic development, it is a priority to increase the scale and frequency of aquatic biodiversity monitoring.
This study also demonstrates that when comparing eDNA metabarcoding with traditional sampling methods on a local scale (a few kilometers), eDNA samples produced a more comprehensive species list. eDNA metabarcoding has been recognized as a more sensitive method than electrofishing or netting.
Results were published and can be accessed here.
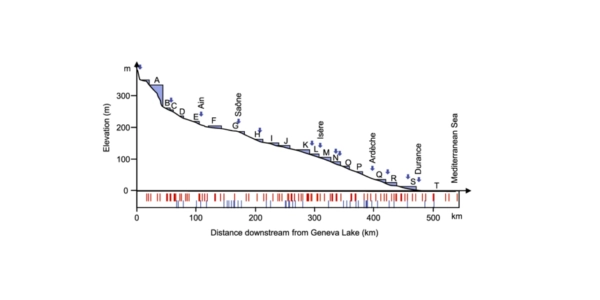
Location: Rhône River from Lake Geneva to the Mediterranean Sea (540 km).
Ecosystem: Freshwater River.
Depth: Surface Sampling.
Sampling method: via two 30 liters filters on 20 successive sections of the Rhône River, using 2 peristaltic pumps on each side of a ship (surface).
Taxonomic group: fishes, using teleo primer.

