
How to detect all species of a target group from an environmental sample?
The VigiDNA M technologies, based on the “eDNA metabarcoding” approach, are mainly used for the monitoring of key taxonomic groups in aquatic ecosystems, for mamal species monitoring from faeces, for the diet analysis of animal species from faeces and for flowering plants composition assessment from honey.


Monitoring of target taxonomic groups in aquatic ecosystems
This approach has been developed since 2011 by SPYGEN and its partners. It is based on the use of universal primer pairs* and Next Generation Sequencing technologies. This non-invasive method enables blind detection of all species of a target group present on the study site and thus represents a powerful environmental monitoring tool. It enables improved detection of rare species (in comparison with conventional methods), a reduction in survey costs and the avoidance of any risk of introduction of pathogens or invasive species during sampling.
For more information, you can download: Valentini et al. 2016.pdf
*Patented technology (CNRS – Université Grenoble 1)

Diet analysis from faecal samples
This non invasive method was developed in 2007 by the Laboratoire d’Ecologie Alpine in order to improve our knowledge on the biology of threatened animal species and their interactions with the ecosystem. It is based on the extraction of DNA from faecal samples and its amplification using an universal primer pair*. The amplified DNA is then sequenced (Next Generation Sequencing) and the obtained sequences are compared to the GenBank international reference database using bioinformatics tools.
For more information, you can download: Soininen et al. 2009.pdf
*Patented technology (CNRS – Université Grenoble 1)

Plant composition from a honey sample
Developed in 2010 by the Laboratoire d’Ecologie Alpine, this method is based on the use of an universal primer pair for plants* and Next Generation Sequencing technologies. Using bees as “environmental samplers”, it is possible to assess the plant biodiversity in a study site and to follow its evolution through time, by regularly analysing honey samples. It also represent a powerful tool for the assessment of honey geographical origin.
For more information, you can download: Valentini et al. 2010.pdf
*Patented technology (CNRS – Université Grenoble 1)
Analyses protocols
-
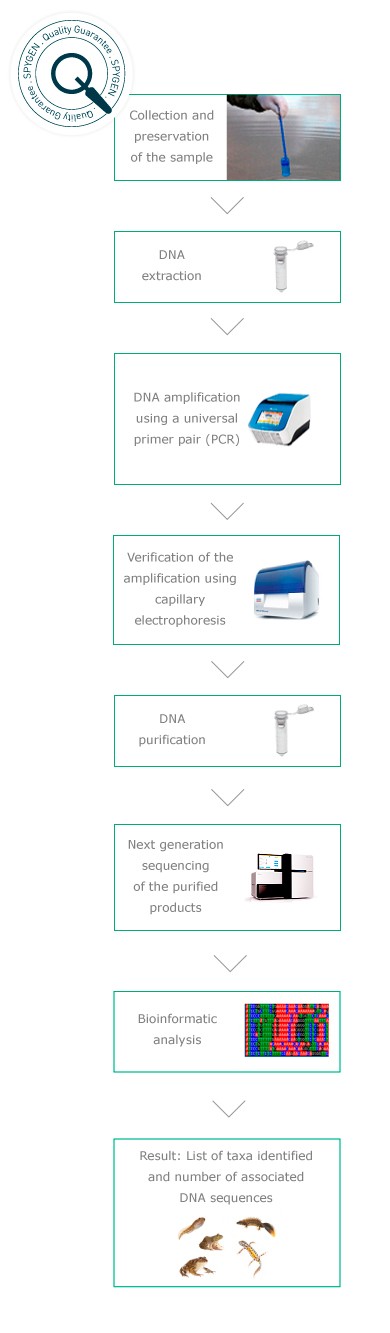
Quality controls
In order to guarantee the quality of our analyses, controls are performed at each step of the protocol. They enable assessment of the DNA extraction yield, verification of the purity of the consumables used, detection of potential cross-contaminations during the analysis or the presence of false positives related to amplification or sequencing errors. All universal primer pairs used in the PCR are validated in silico (bioinformatic analyses), in vitro (from tissue samples) and in situ (on sites where the list of the species present is known). The sequences obtained using Next Generation Sequencing are then compared to reference genetic databases developed specifically by SPYGEN.
-

Quality controls
In order to guarantee the quality of our analyses, controls are performed at each step of the protocol. They enable assessment of the DNA extraction yield, verification of the purity of the consumables used, detection of potential cross-contaminations during the analysis or the presence of false positives related to amplification or sequencing errors. The sequences obtained using Next Generation Sequencing are then compared to the GenBank international reference database.
-
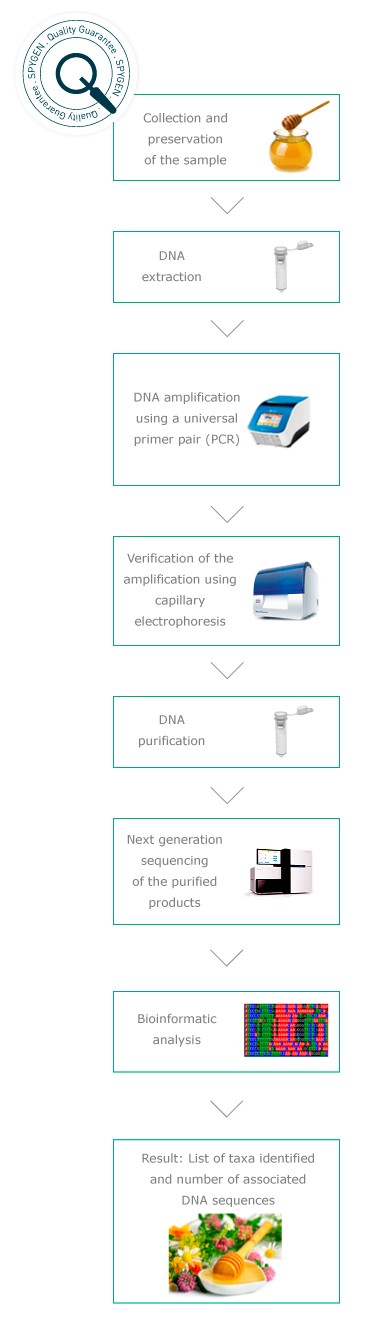
Quality controls
In order to guarantee the quality of our analyses, controls are performed at each step of the protocol. They enable assessment of the DNA extraction yield, verification of the purity of the consumables used, detection of potential cross-contaminations during the analysis or the presence of false positives related to amplification or sequencing errors. The sequences obtained using Next Generation Sequencing are then compared to the GenBank international reference database.

